This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
The RB1 Gene

The RB1 gene is located on the thirteenth chromosome, specifically, 13q14.2. The gene is 114142980 base long.
FASTA Genomic Sequence
Figure 1
Homology
Seven Homologs were found for the RB1 gene:
Pan troglodytes (Chimpanzee)
Canis lupis familiaras (Dog)
Rattus Norvegicus (Norway Rat)
Mus Musculus (Mouse)
Bos Taurus (Cattle)
Gallus Gallus (Chicken)
Danio rerio (Zebra Fish)
Click the links above for the FASTA sequences.
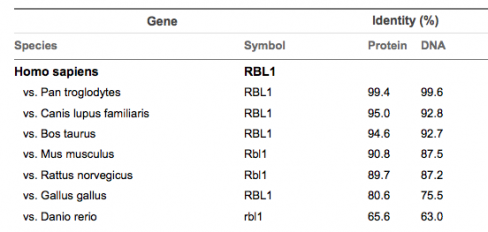
Sequence Similarity
Figure 2
Multiple Alignment
Multiple alignment was done using GeneBee.
| rb1homologs.jpg | |
| File Size: | 282 kb |
| File Type: | jpg |
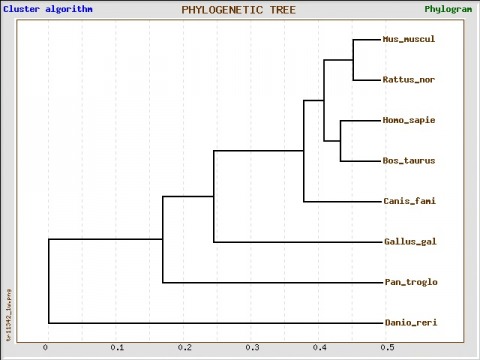
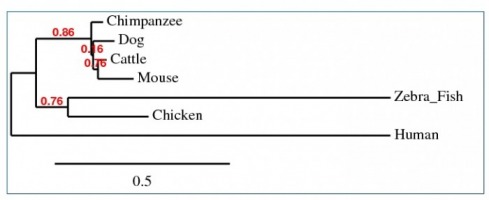
Phylogeny
Phylogenetic tree was constructed with GeneBee, using the seven known homologs of the human RB1 gene. The most closely related to Humans was the Bos Taurus (Cattle). Danio rerio (Zebra Fish) is the least related to the human RB1 gene. These results are odd and were unexpected.
When looking at percent sequence similarities in the above chart, it suggests that the chimpanzee is the most closely related of the homologs, in both protein sequence and DNA sequence. The below tree was constructed using protein sequence information. DNA was also tried and similarly confusing results were discovered as well. The potential source of error is believed to be the result of not being able to use the full sequence of either the protein or the gene. This was due to the limitations of the alignment software used. Several different portions of the sequences were used in an attempt to see if the results were incorrect. No tree was found to match the findings that were found through sequence similarity.
DNA Motifs
Two DNA Motifs were discovered in the Human RB1 gene. The first was discovered in C. Elegans. The gene is SKN1 gene is is required for embryonic endodermal and mesodermal specification, and for maintaining differentiated intestinal cells postembryonically. The second gene motif that was discovered was in Drosophilia, Heat Shock Factor (HSF). HSF is responsible for transcriptionally activating Heat Shock genes, which help cells cope with abnormally high temperatures that have the potential to damage the cell.
References
1. GeneBee http://www.genebee.msu.su/
2. ClustalW http://www.ebi.ac.uk/Tools/clustalw2/index.html
3. NCBI http://www.ncbi.nlm.nih.gov/
4. Figure 1 http://www.davidson.edu/academic/psychology/ramirezsite/neuroscience/psy324/memajure/home.html
5. Homologene http://www.ncbi.nlm.nih.gov/homologene/
6. MOTIF http://motif.genome.jp/
7.Knudson 1971, Proc Acad Nat Sci USA 68:820
8. Figure 2: Homologene, http://www.ncbi.nlm.nih.gov/homologene/
9. Figure 3, 4: GeneBee, http://www.genebee.msu.su/
10. Figure 5: Phylogeny fr, http://www.phylogeny.fr
Justin Lengfeld
[email protected]
Genetics 677
Last Update: 5/13/09